News
Notes about the latest events
02.05.2025 - New preprint: Exploring the Feasibility of AI-Assisted Spine MRI Protocol Optimization Using DICOM Image Metadata
Our new preprint, Exploring the Feasibility of AI-Assisted Spine MRI Protocol Optimization Using DICOM Image Metadata, is now available on arXiv! Developed in collaboration with the Radiology and Medical Physics Services of Hospital de Clínicas de Porto Alegre (HCPA), this study -- part of Alice Vian’s master's research -- investigates how AI can assist in optimizing MRI protocols. By analyzing DICOM metadata, we identified key acquisition parameters that influence image quality, providing valuable insights for medical physicists in quality control and protocol refinement. Read more about our work here.
12.04.2024 - Paper presented at the BSB 2024
Our paper, Evaluating the Generalization of Neural Network-Based Pan-Cancer Classification Models for Cohort-Specific Predictions was presented at the Brazilian Symposium on Bioinformatics (BSB 2024), taking place in Vitória, ES, from December 2–4, 2024. This work, developed as part of Thomas Fontanari’s master’s research, explores the generalization of pan-cancer models using neural networks, including a few-shot learning approach to improve predictions across diverse cohort.
Paper abstract: This study develops and evaluates pan-cancer (PC) models for cohort-specific (CS) predictions using neural networks (NNs). We adopt a dual approach, including a method inspired by few-shot learning, aiming at improving the models’ ability to distinguish between normal and tumorous tissues across diverse cohorts. The first approach trains a NN with comprehensive PC datasets containing 16 cancer types, comparing it against CS models on a target cohort, while the second analyzes whether PC models could generalize to smaller and unseen cohorts by training on 15 cohorts and evaluating on the excluded cohort. Our experiments show that PC models generally outperform CS models, even with limited sample sizes and class imbalances. Moreover, the few-shot approach successfully generalizes to other cancer types, highlighting its potential to advance personalized cancer diagnosis and treatment.
The full paper can be acessed here.
11.30.2024 - New preprint: GNNs for Heart Failure Prediction on an EHR-Based Patient Similarity Graph
New Preprint Alert! Our latest research, Graph Neural Networks for Heart Failure Prediction on an EHR-Based Patient Similarity Graph, is now available on arXiv! This study is part of Heloisa Boll master's work and was developed in collaboration with researchers from Halmstad University. The paper explores the use of Graph Neural Networks (GNNs) and a Graph Transformer (GT) to predict heart failure incidence based on patient similarity graphs built from electronic health records (EHR). Our findings highlight the potential of graph-based models in capturing complex patient relationships, improving both prediction accuracy and interpretability. Check it out in this link.
10.04.2024 - Short paper accepted for STIL 2024
Our paper Beyond Single Models: Leveraging LLM Ensembles for Human Value Detection in Text, developed by Diego Dimer Rodrigues in close collaboration with Profa Viviane Moreira, was accepted as a short paper for presentation in STIL (Symposium in Information and Human Language Technology) 2024. This is the most important Brazilian event for researchers interested in Natural Language Processing (NLP) and it is collocated with BRACIS 2024 (The 34th Brazilian Conference on Intelligent Systems), whicil will take place in Belém, PA.
Paper abstract: Every text might contain its writer's opinion, and every opinion, primarily political, can be associated with the human values it attains or constrains. Identifying those values in the text through machine learning can empower policymakers with a more comprehensive understanding of underlying factors when formulating policies. Current large language models have been employed for various tasks, but one model might not generalize enough to achieve high performance in tasks like value detection. This work aims to leverage multiple ensembles of LLMs to identify human value in text and improve the performance of each model through the ensembles. Our results show that the ensemble models had higher F1 scores than all the baseline models, and the combination of models can be a powerful asset that could perform similarly to the very large models that would require much more memory to solve this task
05.21.2024 - Paper accepted for presentation in IEEE CIBCB 2024
We had a paper accepted for oral presentation in the 21st IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (IEEE CIBCB 2024), which will take place in Natal/RN, Brazil. The paper Predicting Cancer Driver Genes: Leveraging Graph Convolutional Networks and Ensemble Learning brings research results from my student Renan Andrades, who completed his master's degree (PPGC/INF/UFRGS) in 2023.
Paper abstract: Cancer driver genes (CDGs) are crucial in cancer development and are key targets for treatment. This study proposes a novel approach utilizing Graph Neural Networks (GNNs), particularly Graph Convolutional Networks (GCNs), to predict CDGs by integrating protein-protein interaction (PPI) networks and multi-omics data. We optimized GCN hyperparameters across six different PPI networks from literature, which improved predictive accuracy. A consensus model was created using genes identified across all networks. We used ensemble-based methods, including majority voting and score aggregation, to identify high-probability CDGs. Comparisons showed that ensemble models outperformed individual models. Our approach identified several candidate genes, including some previously unclassified, as potential CDGs in various cancers. Notably, 33 genes were consistently predicted as CDGs across all networks, highlighting their potential importance in cancer development. This study highlights the effectiveness of GNNs for CDG prediction and the value of integrating multi-omics data and diverse PPI networks for advancing cancer genomics research and targeted therapy development.
04.18.2024 - New paper published in the journal Biomedical Signal Processing and Control
Excited to share the publication of a new paper in the Biomedical Signal Processing and Control journal, which presents innovative research on using smartphone apps to monitor Aedes aegypti mosquitoes through acoustic analysis using machine learning: Acoustic identification of Ae. aegypti mosquitoes using smartphone apps and residual convolutional neural networks. This study was developed in collaboration with Kayuã Oleques Paim (PhD student, PPGC/INF-UFRGS), Ricardo Rohweder (PPGBM/UFRGS) and professors Rodrigo Brandão Mansilha (PPGES/UNIPAMPA) and Weverton Cordeiro (PPGC/INF-UFRGS). We present a novel residual convolutional neural network (CNN) architecture that classifies mosquito wingbeat sounds recorded on smartphones, focusing on creating a cost-effective and efficient monitoring tool. Our paper introduces a benchmark dataset of mosquito audio recordings and demonstrates the effectiveness of our CNN architecture in real-world conditions by meeting key requirements for robustness to noise and efficient operation on common smartphones. This work highlights the potential for smartphone-based crowd-sourcing solutions to aid in mosquito population control, offering a scalable and accessible tool for public health initiatives.
A preliminary version of this work is also available as a preprint in the arXiv repository.
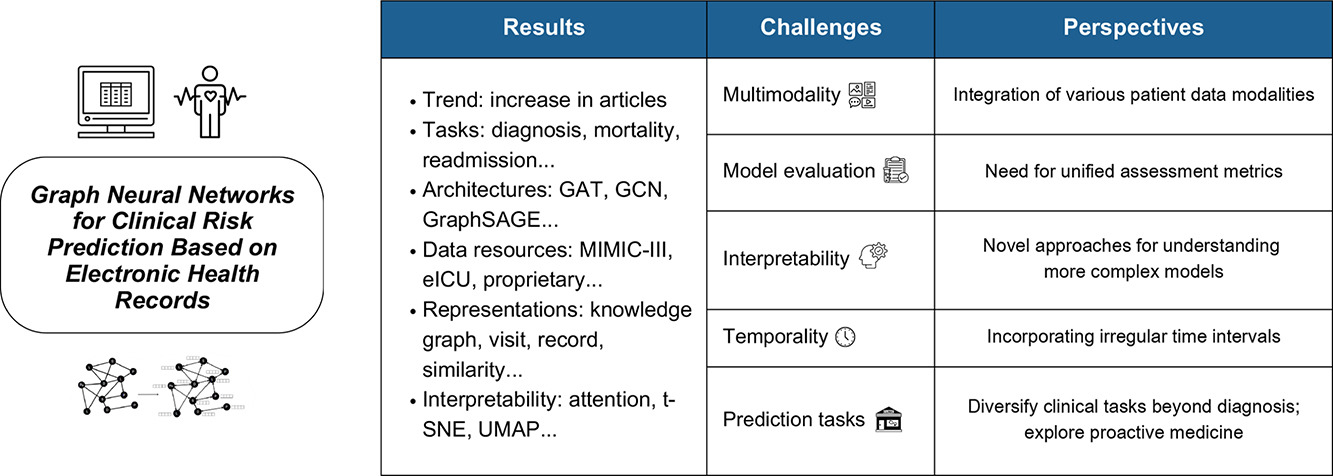
02.23.2024 - New survey paper about GNNs for clinical risk prediction using EHRs
Thrilled to announce the acceptance of the paper Graph Neural Networks for Clinical Risk Prediction Based on Electronic Health Records: A Survey, authored by my Master's student, Heloísa Oss Boll, and collaborators from Brazil (Institute of Informatics, UFRGS) and Sweden (School of Information Technology, Halmstad University), in the Journal of Biomedical Informatics. This paper provides a comprehensive review of the use of graph neural networks (GNNs) for clinical risk prediction using electronic health records (EHRs), highlighting the growing research interest and identifying key challenges in this area. It examines 50 relevant articles, noting the prevalence of graph attention networks (GAT) and the MIMIC-III dataset, and suggests future directions to improve model accuracy and applicability in clinical settings. The full text of the paper is freely available in this link.
02.16.2024 - New paper! NBioinfo: Establishing a Bioinformatics Core in a University-based General Hospital in South Brazil
We had a paper published in a special edition of the Journal of Information and Data Management (Vol. 15, No. 1, 2024), that aims to publicize research groups in bioinformatics in Brazil. Our paper NBioinfo: Establishing a Bioinformatics Core in a University-based General Hospital in South Brazil shares the history of the Núcleo de Bioinformática (NBioinfo) at the Hospital de Clínicas de Porto Alegre (HCPA) in Brazil. This initiative aims to address the critical need for bioinformatics expertise in tackling the growing volume and complexity of biological data in life and medical sciences. Since its inception, NBioinfo has served as a vital hub for research, collaboration, and education in Bioinformatics and Computational Biology, fostering institutional development and scientific advancement. The paper highlights the challenges and successes in building this interdisciplinary field at HCPA, emphasizing the importance of bioinformatics in enhancing biomedical research and healthcare outcomes. The special edition was organized by the Special Committee on Computational Biology (CE-BioComp) of the Brazilian Computing Society (SBC).
01.20.2024 - On maternity leave for the next six months!
I'm happy to share the joyful news that I will be taking a maternity leave to welcome our new addition to the family - Helena. My daughter was born on January 10th, 2024. During this special time, I will be taking some time away from professional activities to focus on my family and enjoy this precious journey. I'll return from maternity leave (+ vacation) in the end of July.
12.23.2023 - Two papers accepted for publication in Genetics and Molecular Biology
We had two papers accepted for publication in a special issue of the journal Genetics and Molecular Biology, organized for the celebration of the 60 years of the Graduate Program in Genetics and Molecular Biology (PPGBM) of UFRGS.
The paper "Broken silence: 22,841 predicted deleterious synonymous variants identified in the human exome through computational analysis", with Ana Carolina Mello as first author, explores the role of synonymous single nucleotide variants (sSNVs) in human exomes and their potential deleterious effects. The work aims to develop a framework to assist in the deleteriousness prediction of sSNVs identified by whole exome sequencing (WES) data, using heterogeneous ensemble feature selection. The research was supervised by Ursula Matte, and developed in collaboration with Felipe Colombelli, Delva Leão, Luis Dias, and Andreia Carina Turchetto-Zolet.
The paper Brazilian women in Bioinformatics: Challenges and opportunities aims to discuss the pivotal role of women in the rapidly growing field of bioinformatics, especially highlighted during the COVID-19 pandemic, and challenges women encounter, such as prejudice and stereotypes. The paper delves into the historical context of bioinformatics, emphasizing its applications in genetics and the gender disparities women face in STEM fields. The work was developed in collaboration with Thayne Woycinck Kowalski, Giovanna Câmara Giudicelli, Maria Clara de Freitas Pinho, Marília Körbes Rockenbach, Miriãn Ferrão Maciel-Fiuza, and Fernanda Sales Luiz Vianna.
12.15.2023 - Concluding the Project 'Building Citizen Science Intelligence for Pandemic Preparedness and Response: Pilot Implementation'
This month we concluded the activities of the project Building Citizen Science Intelligence for Pandemic Preparedness and Response: Pilot Implementation, which aimed to investigate and seek to validate that participatory modelling approaches are usable and useful for pandemic preparedness and response across different levels (i.e., individual, organisational, and community). In this pilot study, conducted in parallel in three countries (Vietnam, Kenya and Brazil), we engaged with specific communities to co-create and roll out participatory modelling artefacts within the specific problem domain of exploring the impact of a pandemic scenario, such as COVID-19, on the management of resources in healthcare facilities and their surrounding communities.
In Brazil, the project was carried out at HCPA under my coordination. The other co-PIs are Dra. Sylvia Muyingo (APHRC, Kenya) and Dr. Marc Choisy (OCRU, Vietnam). The general coordinator of the project is Dr. Serge Stinckwich (United Nations University Institute in Macau). The project was financed by HealthAI, a global, Geneva-based nonprofit organization focused on responsible AI in the health sector.
10.20.2023 - Research Topic and Editorial: 'Bioinformatics Applied to Neuroscience'
Together with Dra. Thayne Kowalski, Dra. Clévia Rosset, and Dra. Jaqueline Bohrer Schuch, we organized a Frontiers Research Topic titled "Bioinformatics Applied to Neuroscience". This Research Topic contains 15 articles presenting current bioinformatics approaches applied to understanding the biological underpinnings of psychiatric and neurological conditions. We also published an editorial, summarizing the contributions of all the original articles included in our collection. Our reseach topic has already received more than 40k views and had almost 15k paper downloads.
10.05.2023 - Published book chapter with an overview of AI, its applications and challenges (in portuguese)
The Graduate Program in Computer Science (PPGC) of the Federal University of Rio Grande do Sul (UFRGS) held on the 5th and 6th of October the "School of Computing – PPGC/UFRGS 50 Years" as part of the celebration of PPGC’s 50th anniversary. The school featured eight presentations that addressed cutting-edge topics in various areas of research at PPGC. Each presentation was accompanied by a book chapter, contributing to the dissemination of knowledge generated at the event. I contributed to a chapter written among many hands that aims to provide an overview of the field of Artificial Intelligence, its applications, opportunities and challenges. The chapter "A Nova Eletricidade": Aplicações, Riscos e Tendências da IA Moderna (in portuguese) is available online as part of the book (978-85-7669-558-5) and also as a preprint. Hope you enjoy it!
Abstract: The provocative comparison between AI and electricity, made by computer scientist and entrepreneur Andrew Ng, summarizes the deep transformation that recent advances inArtificial Intelligence (AI) have triggered in the world. This chapter provides an over-view of the ever-evolving landscape of AI. Without intending to exhaust the subject, we explore the applications that are redefining sectors of the economy, impacting society andhumanity. We analyze the risks that accompany rapid technological progress and future trends in AI, an area that is on the path to becoming a general-purpose technology, just like electricity, which revolutionized society in the 19th and 20th centuries.
09.30.2023 - Papers presentation at BRACIS 2023
From the 25th to the 29th of September I participated in the 12th Brazilian Conference on Intelligent Systems (BRACIS 2023), which was held in Belo Horizonte - MG, on the UFMG campus. This year we had two papers accepted for oral presentation.
The paper Prediction of Cancer-Related miRNA Targets Using an Integrative Heterogeneous Graph Neural Network-Based Method describes results from the Master Thesis of my student Emanoel Vianna, defended in April 2023. We tackle the challenges of imbalanced classes and false positives in predicting miRNA-target interactions by proposing a Graph Neural Network (GNN)-based model using the HinSAGE algorithm, which integrates miRNA-mRNA and mRNA-mRNA interactions with cancer-related gene expression data.
The second paper, Explainability of COVID-19 Classification Models Using Dimensionality Reduction of SHAP Values, is first authored by Daniel Kuhn, who was supervised by Profa Viviane Moreira. This study evaluates multiple machine learning classifiers to predict COVID-19 mortality risk using data available at patient admission, highlighting the importance of predictive models during the pandemic. By employing a visualization technique with a state-of-the-art explainability approach and dimensionality reduction, our work provides insights into the features influencing classifier predictions, achieving a sensitivity of up to 84% and an AUROC of 92% on two real datasets.
09.13.2023 - Paper published in the journal Thyroid
Our paper Classification of Thyroid Tumors Based on DNA Methylation Patterns, developed in collaboration with Vicente Rodrigues Marczyk, Dra. Ana Luiza Maia, and Dr. Iuri Martin Goemann from UFRGS/HCPA, was published in the journal Tyhroid. Using an unsupervised machine learning approach, we analyzed 810 thyroid samples to identify distinct methylation subtypes without prior clinical or pathological data. Our approach successfully classified the samples into three methylation subtypes—normal-like, follicular-like, and papillary thyroid carcinoma (PTC)-like—demonstrating a strong association with histological diagnoses and genomic drivers. Notably, the results highlight the heterogeneity within follicular variant PTC (FVPTC), showing its division into two subtypes linked to different mutations. Our findings underscore the potential of DNA methylation as a biomarker for more precise classification and understanding of thyroid neoplasms.
08.20.2023 - Chapter in the e-book 'Real-life healthcare data in Latin America: trials and insights'
Roche Farma has launched an e-book coordinated by PEV Consultoria em Saúde and developed with experts in the field to promote education and awareness about the use of real-world data for various health purposes. This initiative reflects the digital transformation in healthcare, emphasizing the potential of real-world data to inform strategies that positively impact public health. I contributed to the chapter Brazilian health databases as a source of real-world data, written in collaboration with MSc Miriam Allein Zago Marcolino, Dra. Ana Paula Beck da Silva Etges, and Dra. Carisi Anne Polanczyk.
The e-book Dados de vida real em saúde na América Latina: ensaios e percepções is available online (in portuguese only).
08.25.2022 - Paper accepted for publication in BRACIS 2022
Our paper Cross-validation Strategies for Balanced and Imbalanced Datasets has been accepted for publication in the 11th Brazilian Conference on Intelligent Systems (BRACIS 2022)! This work was developed by my master student Thomas Fontanari and my undergraduate student Tiago Froes. BRACIS is one of the most important events in Brazil for researchers interested in Artificial and Computational Intelligence.
Paper abstract: Cross-validation (CV) is a widely used technique in machine learning pipelines. However, some of its drawbacks were recognized in the last decades. In particular, CV may generate folds unrepresentative of the whole dataset, which led some works to propose methods that attempt to produce more distribution-balanced folds. In this work, we propose an adaption of a cluster-based technique for cross-validation based on mini-batch k-means that is more computationally efficient. Furthermore, we compare our adaptation with other splitting strategies previously not compared and also analyze whether class imbalance may influence the quality of the estimators. Our results indicate that the more elaborate CV strategies show potential gains when a small number of folds is used, but stratified cross-validation is preferable for 10-fold CV or in imbalanced scenarios. Finally, our adaptation of the cluster-based splitter reduce its computational cost while retaining similar performance.
08.23.2022 - Paper published in Knowledge-Based Systems
The paper A hybrid ensemble feature selection design for candidate biomarkers discovery from transcriptome profiles, produced during the scientific initiation activities of my undergraduate student Felipe Colombelli, has been accepted for publication in the journal Knowledge-Based Systems! Our feature selection method explores perturbation in data and methods level to disrupt associations of good performance with a single dataset, single algorithm, or a specific combination of both, which is particularly interesting for better reproducibility of genomic biomarkers. In this paper, we explored cancer-related transcriptome, but the method is applicable to other types of omics. Interestingly, the ranks produced by our method reached greater biological plausibility than other tested approaches, with a notably high enrichment for cancer-related genes and pathways. We had an important collaboration of Dra. Thayne Kowalski for the bioinformatics analysis and interpretation of biological results. The project has received partial funding from CNPq and FAPERGS. The developed tools were made freely available: efs-assembler, a Python package implementing various functionalities for ensemble feature selection analysis, and BioSelector, a graphical user interface solution written using Electron/React (widely used desktop front-end solutions in industry) to support multiple operating systems and a user- friendly experience.
08.15.2022 - Paper accepted for the Full Paper Track at BSB2022
Our paper Study on the complexity of omics data: an analysis for cancer survival prediction, authored by my undergraduate student Carlos Daniel Andrade (CIC/INF) and my master student Thomas Fontanari (PPGC/INF), was accepted for presentation in the Full Paper Track of the 15th Brazilian Symposium on Bioinformatics (BSB 2022). BSB is an international conference which covers all aspects of Bioinformatics and Computational Biology, and is organized by the special interest group in Computational Biology (CE-BioComp) of the Brazilian Computer Society (SBC)
Paper abstract: The use of machine learning approaches in studying cancer through omics datasets has been an important research tool since the advent of high-throughput technologies. However, these datasets present an intrinsic data complexity that may hinder model development despite their information richness. This work, therefore, aims to study the characteristics of different omics data commonly employed for clinical predictive analysis using a broad set of data complexity measures tailored for imbalanced domains. We focus on the task of cancer survival prediction in eight tumor types based on four types of omics data (\ie copy number variation, gene expression, microRNA expression, and DNA methylation) and the combination among them (multi-omics approach). We found that F1-MaxDr, F3 partial, F4 partial, and N3 partial could be used as predictors of performance. Furthermore, our experiments suggested that the studied omics data types are strongly correlated in terms of data complexity, including the multi-omics approach. All eight cancer types appeared to be highly correlated with each other, except for Adrenocortical Carcinoma (ACC), which showed a significantly lower complexity than the others.
07.05.2022 - Two new transcriptome meta-analyses published
In the last weeks, we had two papers accepted and published with results from transcriptome meta-analyses studies. These works were developed in collaboration with Dra. Thayne Kowalski, Dra. Fernanda Vianna, and Dra. Giovanna Giudicelli. In one paper published by European Neuropsychopharmacology, we applied this methodology to investigate the effect of Valproic acid (VPA) - a widely used antiepileptic drug not recommended in pregnancy because it is teratogenic - on gene expression profiles. In the second paper recently accepted by Neuroscience Informatics, the methodology was employed to evaluate the impact of ethanol in the differential gene expression of embryonic cells and fetal tissues, and further study the molecular mechanisms of Fetal Alcohol Spectrum Disorder (FASD).
06.29.2022 - Panel presented at the Global Health Security Conference 2022
From June 28th to July 1st, I attended the Global Health Security Conference (GHSC'22) organized by the Global Health Security Network, held at Singapore. Along with Peiling Yap, chief scientist at I-DAIR, and my colleagues Marc Choisy (Oxford University Clinical Research Unit) and Pey Canlas (Wireless Access for Health, WAH) from the pandemic scientific working group led by I-DAIR, we prepared and presented the panel Citizen science to drive a more responsive and inclusive digitally-enabled pandemic preparedness and response scheme. My talk titled Multidisciplinary and participatory approaches in data collection or more effecti e data-driven responsive pandemic surveillance discussed the importance in broadening data collection for surveillance schemes by enhancing data diversity and better engaging citizens as active actors in this process. It was an enriching experience both to share with GHSC attendees the work we have been developing with I-DAIR and to listen to so many inspiring talks about recent advances and the various remaining challenges to make a world healthier and safer for all.

06.27.2022 - New Paper! A call for citizen science in pandemic preparedness and response
Happy to share that the paper A call for citizen science in pandemic preparedness and response: beyond data collection, led by I-DAIR with collaboration from the members of the Pandemic Scientific Working Group, has been published by BMJ Global Health! Beautiful joint work that brings attention to the potential of digitally enabled citizen science in translating data into accessible, comprehensible and actionable outputs at the population level, particularly in pandemic preparedness and response.
05.15.2022 - Project Mosquitoramento publicized in the media
During the past week, the Mosquitoramento project was publicized in the media at two different times. After being awarded first place at the Unicred Health Alliance Program, our project was discussed in a newspaper article written by Juliana Bublitz for Zero Hora (one of the largest daily newspapers in Brazil), also available at GZH website (digital version).
Morever, on May 12th, we made a live participation in the news "Bom Dia Rio Grande" to talk about our project and how the idea that the Mosquitoramento aims to implement can make the monitoring of Aedes aegypii mosquitoes broader and more agile through artificial intelligence. Check the video here (in portuguese).
05.06.2022 - Project Mosquitoramento awarded 1st prize in the Unicred Porto Alegre Health Alliance
The project Mosquitoramento, coordinated by me and my colleagues Prof. Weverton Cordeiro and Prof. José Rodrigo Azambuja, was the first prize winner of the Unicred Porto Alegre Health Alliance Program! The event happened during the South Summit Brazil Porto Alegre and was created was to stimulate the development of innovative solutions in the health area. For more information, see this link (in portuguese).
03.21.2022 - Survey paper published in Briefings in Bioinformatics
Excited to share that our work Machine learning methods for prediction of cancer driver genes: a survey paper has been published in Briefings in Bioinformatics! In this survey, prepared as part of Renan Andrades's Master Thesis at PPGC/INF/UFRGS, we aimed to summarize the main efforts towards the development of ML models for the task of discovering new cancer drivers. We reviewed a large body of papers and focused on constructing an integrated, panoramic view of data and ML techniques within this domain. We also reflect about several methodological challenges that remain to be explored to accelerate the computational discovery of cancer drivers. Briefings in Bioinformatics is among the top venues for Bioinformatics publications and its current impact factor (JCR 2020) is 11.622!
03.04.2022 - Paper published in Psychiatry Research
The paper Identifying posttraumatic stress disorder staging from clinical and sociodemographic features: a proof-of-concept study using a machine learning approach has been published in the journal Psychiatry Research. This work was developed as part of the PhD thesis of Dr. Luis Francisco Ramos-Lima under the supervision of Profa. Dra. Lucia Helena Machado Freitas, in the Post-Graduate Program in Psychiatry and Behavioral Sciences at UFRGS. I had the pleasure to collaborate with them in divising and implementing the machine learning methodology to develop a predictive model to support posttraumatic stress disorder (PTSD) staging.
01.05.2022 - Research proposal approved by FAPERGS - Programa Pesquisador Gaúcho (PqG)
I had a project approved by the State Research Funding Agency of Rio Grande do Sul (FAPERGS) call for the Programa Pesquisador Gaúcho (PqG)! The project "MARCS: Modelos de Aprendizado de máquina Robustos e Confiáveis para a Saúde" (Robust and Reliable Machine Learning Models for Health) will receive funding during the next three years to develop research that aims to advance our scientific knowledge about strategies to diagnose and mitigate bias problems in ML models applied to Health, contributing to the development of more robust and reliable models, both from a technical and ethical point of view.
01.01.2022 - Fellowship of Research Productivity (PQ) granted by CNPq
Great news to start 2022! The National Council for Scientific and Technological Development (CNPq) has approved my research proposal in the context of the Call CNPq No 04/2021 - Bolsas de Produtividade em Pesquisa (Research Productivity Fellowship), which aims to provide a financial recognition for researchers that excel in their academic and scientific activities. Starting in 2022, I'll be a CNPQ Research Productivity Fellow Level 2 (PQ-2) for a period of three years.
12.20.2021 - Paper published in Genetics and Molecular Biology
The paper Gene Expression Analysis Platform (GEAP): A highly customizable, fast, versatile and ready-to-use microarray analysis platform, co-authored with Itamar Nunes and Bruno César Feltes (Bioscience Institute, UFRGS) was published in Genetics and Molecular Biology. Check its abstract below or read the full text on the journal's website (open access).
There are still numerous challenges to be overcome in microarray data analysis because advanced, state-of-the-art analyses are restricted to programming users. Here we present the Gene Expression Analysis Platform, a versatile, customizable, optimized, and portable software developed for microarray analysis. GEAP was developed in C# for the graphical user interface, data querying, storage, results filtering and dynamic plotting, and R for data processing, quality analysis, and differential expression. Through a new automated system that identifies microarray file formats, retrieves contents, detects file corruption, and solves dependencies, GEAP deals with datasets independently of platform. GEAP covers 32 statistical options, supports quality assessment, differential expression from single and dual-channel experiments, and gene ontology. Users can explore results by different plots and filtering options. Finally, the entire data can be saved and organized through storage features, optimized for memory and data retrieval, with faster performance than R. These features, along with other new options, are not yet present in any microarray analysis software. GEAP accomplishes data analysis in a faster, straightforward, and friendlier way than other similar software, while keeping the flexibility for sophisticated procedures. By developing optimizations, unique customizations and new features, GEAP is destined for both advanced and non-programming users.
GEAP's website is available here.
10.21.2021 - ICCSA 2021 Best Paper Award winner
The paper reporting the results from the research project developed by my student Bernardo Trevizan during his work completion of graduation was selected for the list of Best Paper Award winners of the 21st International Conference on Computational Science and its Applications (ICCSA 2021). Congrats, Bernardo!! Our paper is available online.

10.15.2021 - Our Mosquitoramento team takes 3rd place in the Flex Challenge of Technological Innovation
The team of students from UFRGS Brayan Barbosa (Biomedicine), Eduardo Peretto (Computer Science), Lucas Dal Castel (Electrical Engineering), Luiz Felipe de Moura (Control and Automation Engineering) and Mateus Schein (Control and Automation Engineering), coordinated by professors from the Institute of Informatics José Rodrigo Azambuja, Mariana Mendoza, and Weverton Cordeiro (INA) took third place in the second edition of the Flex Challenge of Technological Innovation. Our Mosquitoramento team presented a low-cost system for detecting Aedes aegypti mosquitoes through audio. The challenge had more than 120 teams registered, which competed over 5 months. In the final phase of the competition, which took place from October 4th to 8th in Sorocaba (SP), in person, the team presented a 7-minute pitch followed by a demonstration of the solution's functionality, which includes multiple clients for recording and sending audio, a remote server running a neural network for audio classification and detection of Aedes aegypti and a dashboard for system control. The panel of evaluators, made up of investors and entrepreneurs, ranked the team in 3rd place, with a cash prize of R$7,000, second only to the projects "A Liconic Power Unit" (FACENS) e "Guara e Irae" (FACENS). Check the news on the UFRGS's page (in portuguese).

10.14.2021 - New preprint available! Machine learning methods for prediction of cancer driver genes: a survey paper
The survey about machine learning methods for the prediction of cancer driver genes, prepared as part of my student Renan Andrades' master's research (PPGC/INF), is available as a preprint in ArXiV.
Identifying the genes and mutations that drive the emergence of tumors is a major step to improve understanding of cancer and identify new directions for disease diagnosis and treatment. Despite the large volume of genomics data, the precise detection of driver mutations and their carrying genes, known as cancer driver genes, from the millions of possible somatic mutations remains a challenge. Computational methods play an increasingly important role in identifying genomic patterns associated with cancer drivers and developing models to predict driver events. Machine learning (ML) has been the engine behind many of these efforts and provides excellent opportunities for tackling remaining gaps in the field. Thus, this survey aims to perform a comprehensive analysis of ML-based computational approaches to identify cancer driver mutations and genes, providing an integrated, panoramic view of the broad data and algorithmic landscape within this scientific problem. We discuss how the interactions among data types and ML algorithms have been explored in previous solutions and outline current analytical limitations that deserve further attention from the scientific community. We hope that by helping readers become more familiar with significant developments in the field brought by ML, we may inspire new researchers to address open problems and advance our knowledge towards cancer driver discovery.
09.11.2021 - Paper accepted for oral presentation at ICCSA 2021
A paper first-authored by Bernardo Trevizan, who concluded his CS undergraduate course in May 2021, was accepted for oral presentation at the 21st International Conference on Computational Science and its Applications (ICCSA 2021). The paper is titled Ensemble Feature Selection Compares to Meta-analysis for Breast Cancer Biomarker Identification from Microarray Data. It describes the results of an homogeneous ensemble feature selection approach that leverages seven microarray datasets to identify subsets of genes that are stable and show good predictive power for breast cancer, thus representing candidate biomarkers for the disease. The classifiers were tested on six independent test sets. We observed that the results were very competitive against the Random Effect Model meta-analysis, a state-of-the-art approach for microarray integrative analysis. The conference proceedings were published in Springer in the Lecture Notes in Computer Science series, and are available online.
08.16.2021 - Paper published in Computers in Biology and Medicine
Our paper Patterns of high-risk drinking among medical students: a web-based survey with machine learning was recently published in Computers in Biology and Medicine! This paper results from a collaborative work with Prof. Dr. Ives Passos' group, from Department of Psychiatry at UFRGS and Laboratory of Molecular Psychiatry at HCPA, and was mainly developed during the undergraduate thesis of Flávia Pereira (CS student) with close partnership with MD Grasiela Marcon - who shared co-first authorship of this work. The research had important contributions from Prof. Dr. Bruno Castro da Silva (INF/UFRGS) and Profa. Dra. Lisia von Diemen (Department of Psychiatry/UFRGS).
08.01.2021 - New preprint available: A Hybrid Ensemble Feature Selection Design for Candidate Biomarkers Discovery from Transcriptome Profiles
The results of the work developed by Felipe Colombelli (CS undergraduate student) during his scientific initiation activities were recently published as a preprint. In this paper, we proposed a hybrid ensemble feature selection approach to identify potential biomarkers from high-dimensional datasets such as transcriptomic data, also performing an extensive comparison with other traditional and ensemble approaches. Moreover, we have developed two open-source tools to support further studies with similar goals: BioSelector, a user-friendly application, and efs-assembler, a plain Python package. We would be happy to hear feedback about these tools in case you have the chance to use them in your research!
05.26.2021 - Abstract accepted for presentation at ISMB/ECCB 2021
Our abstract describing the organization and the results of the Bioin4Girls: Brazilian (women) symposium in Bioinformatics, an online event promoted in 2020 by the Bioinformatics Core of Hospital de Clínicas de Porto Alegre (HCPA), has been accepted for poster presentation at ISMB/ECCB 2021, in the track Education COSI: Computational Biology Education. BioIn4Girls was an online event with talks spanning seven major topics in bioinformatics, all given by female researchers. Besides, 100% of the steering committee members were women. The ISMB/ECCB conference will take place in July 2021 in a completely virtual format.
05.18.2021 - Paper accepted for publication in Frontiers in Genetics
The paper Comparative Genomics Identifies Putative Interspecies Mechanisms Underlying Crbn-Sall4 Linked Thalidomide Embryopathy has been accepted for publication in Frontiers in Genetics, section Toxicogenomics. This work has shared co-first authorship by Thayne Woycinck Kowalski and Gabriela Barreto Caldas-Garcia, and is a collaboration among researchers from Universidade Federal do Rio Grande do Sul (UFRGS), Hospital de Clínicas de Porto Alegre (HCPA), and Universidade Federal da Bahia (UFBA).
02.21.2021 - Paper published in Obesity Surgery
The paper Roux-en-Y Gastric Bypass Downregulates Angiotensin-Converting Enzyme 2 (ACE2) Gene Expression in Subcutaneous White Adipose Tissue: A Putative Protective Mechanism Against Severe COVID-19, describing results of a collaboration with researchers from Hospital de Clínicas de Porto Alegre (HCPA) and Virginia Commonwealth University, was published in Obesity Surgery. In this work, by reanalyzing publicly available data, we found that angiotensin-converting enzyme 2 (ACE2) gene expression is downregulated in subcutaneous white adipose tissue after the Roux-en-Y Gastric Bypass (RYGB) surgery. Considering that ACE2 is the receptor for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), our results indicate that RYGB may be a potential protective mechanism against severe COVID-19 in obese patients.
01.28.2021 - Paper accepted for publication in IEEE/ACM Transactions on Computational Biology and Bioinformatics
The results of the research conducted by João Shapke (CIC/INF/UFRGS) as a scientific initation student have been accepted for publication in IEEE/ACM Transactions on Computational Biology and Bioinformatics! The paper EPGAT: Gene Essentiality Prediction With Graph Attention Networks, previously made available as a preprint, is already accessible in the "Early Access" area on IEEE Xplore. This work was supervised by me and Prof. Dr. Anderson Tavares. João was a recipient of a CNPq Scientific Initiation Scholarship.
11.25.2020 - Paper accepted for publication in Computers in Biology and Medicine
Our work on the development of deep learning-based classifiers for the detection of Aedes aegypti using audio classification, previously presented as a preprint at Arxiv, was accepted for publication in Computers in Biology and Medicine. The paper is titled Detecting Aedes aegypti Mosquitoes through Audio Classification with Convolutional Neural Networks and describes three strategies - a binary, a multiclass, and an ensemble classifier - using convolutional neural networks to identify Aedes aegypti based on the analysis of recordings from mosquitoes wingbeats captured with smartphones. This work is a proof-of-concept towards the development of an easy-to-use solution that can be widely used for crowd-sourcing the mapping of Aedes aegypti.
This work was developed as the undergraduate thesis of Marcelo Schreiber Fernandes in Electrical Engineering/UFRGS and was co-supervised by prof. Weverton Cordeiro.
11.16.2020 - Paper accepted for publication in Frontiers in Neuroscience
Our paper Anticonvulsants and Chromatin-Genes Expression: A Systems Biology Investigation, resulting from a collaboration with Dra. Thayne Woycinck Kowalski, has been accepted for publication in Frontiers in Neuroscience! This paper evaluates the expression of epigenetics-related genes in valproic acid, carbamazepine, or phenytoin exposure, aiming at identifying drug-induced damage to the embryofetal development. Through bioinformatics analyses, we identified the enrichment of chromatin remodeling genes for the three drugs and a set of candidate genes as potential biomarkers that could be further explored for developing strategies for the prevention of congenital anomalies.
It is important to note that this article is authored only by women: Thayne Woycinck Kowalski, Julia do Amaral Gomes, Mariléa Furtado Feira, Ágata de Vargas Dupont, Mariana Recamonde-Mendoza, and Fernanda Sales Luiz Vianna! :-)
10.22.2020 - Invitation to participate in the 11th BRAGFOST Symposium
I'm very happy and honored to be one of the Brazilian reseachers selected as a participant of the 11th Brazilian-Germany Frontier of Science and Technology Symposium (BRAGFOST 2020), promoted by CAPES and the Alexander von Humboldt Foundation. BRAGFOST 2020 will happen on October 22-23, in an online format due to the current pandemic.
BRAGFOST aims at bringing together outstanding young researchers (up to 15 years after PhD) from Brazil and Germany, who are selected by the organizers based on their contributions and expertise in the research topics covered by the event. This year, the event will have three sessions: Personalized Medicine (for which I was invited), Augmented Intelligence and Education, Biotechnology for Future Sustainability. The main purpose of the event is to stimulate individual exchange between all attendees to foster new research initiatives, resulting in joint projects among Brazilian and German researchers.
More details about the event and the Personalized Medicine session may be found here (portuguese only).
10.01.2020 - BioIn4Girls: Series of Talks by Women in Bioinformatics
The Bioinformatics Core of the Hospital de Clínicas de Porto Alegre (HCPA) is organizing the event BioIn4Girls, a series of talks in Bioinformatics that aims to disseminate all the research possibilities that Bioinformatics offers, which have gained importance worldwide by its relatively low cost and innovative potential. But, more than that, the central goal of the event is to value the work and enhance women's role in this research area through the organization of a program where 100% of the speakers are (fantastic) women. Our goal is to show the importance of female scientists in conducting and leading research in the areas of Bioinformatics and Computational Biology.
The talks will address several research lines in Bioinformatics - Introduction to Bioinformatics, Omics Sciences, Evolution, Metagenomics, Structural Bioinformatics, Analysis of Variants, Machine Learning with Biological Data - applied to different domains, as well as the themes Entrepreneurship and Women in Science. The event will be online and free, every Tuesday and Thursday from October 6th to November 5th, between 6:30pm and 8:30pm. A certificate will be issued for participants with a minimum frequency of 75%.
For more information about speakers, organization, and registration, visit BioIn4Girls' Instagram.
We thank the support of: Hospital de Clínicas de Porto Alegre (HCPA); Institute of Informatics (INF), UFRGS; PPG in Computer Science (PPGC), UFRGS; PPG in Medicine: Medical Sciences (PPGCM), UFRGS; and PPG in Genetics and Molecular Biology (PPGBM), UFRGS

09.27.2020 - Paper accepted by Molecular Diagnosis & Therapy
The article describing the results of the research developed by Rodrigo Haas Bueno during his final undergraduate work in Biomedicine (UFRGS) was accepted for publication by Molecular Diagnosis & Therapy. The paper is titled Meta-analysis of Transcriptomic Data Reveals Pathophysiological Modules Involved with Atrial Fibrillation and the preprint is available at medRXiv.
09.25.2020 - Research project approved by CNPq/AWS
Our project Diagnóstico e predição clínica em oncologia: análise integrativa de dados ômicos através de modelos complexos de aprendizado de máquina (Diagnosis and clinical prediction in oncology: integrative analysis of omic data through complex machine learning models) was one of the proposals selected by CNPq/AWS for receiving Cloud Credits for Research. This project was written in collaboration with Dr. Iuri Martin Goemman (HCPA) and Profa. Dra. Ana Luiza Maia (UFRGS/HCPA) and aims at developing machine learning-based solutions to improve the quality and stability of candidate biomarkers for breast and thyroid cancer.
08.19.2020 - New preprint available: Detecting Aedes Aegypti Mosquitoes through Audio Classification with Convolutional Neural Networks
The preprint Detecting Aedes Aegypti Mosquitoes through Audio Classification with Convolutional Neural Networks is available at ArXiV. This work describes the results of the undergraduate thesis of Marcelo Fernandes (Electrical Engineering/UFRGS), supervised by me and Prof. Dr. Weverton Cordeiro.
The incidence of mosquito-borne diseases is significant in under-developed regions, mostly due to the lack of resources to implement aggressive control measurements against mosquito proliferation. A potential strategy to raise community awareness regarding mosquito proliferation is building a live map of mosquito incidences using smartphone apps and crowdsourcing. In this paper, we explore the possibility of identifying Aedes aegypti mosquitoes using machine learning techniques and audio analysis captured from commercially available smartphones. In summary, we downsampled Aedes aegypti wingbeat recordings and used them to train a convolutional neural network (CNN) through supervised learning. As a feature, we used the recording spectrogram to represent the mosquito wingbeat frequency over time visually. We trained and compared three classifiers: a binary, a multiclass, and an ensemble of binary classifiers. In our evaluation, the binary and ensemble models achieved accuracy of 97.65% (± 0.55) and 94.56% (± 0.77), respectively, whereas the multiclass had an accuracy of 78.12% (± 2.09). The best sensitivity was observed in the ensemble approach (96.82% ± 1.62), followed by the multiclass for the particular case of Aedes aegypti (90.23% ± 3.83) and the binary (88.49% ± 6.68). The binary classifier and the multiclass classifier presented the best balance between precision and recall, with F1-measure close to 90%. Although the ensemble classifier achieved the lowest precision, thus impairing its F1-measure (79.95% ± 2.13), it was the most powerful classifier to detect Aedes aegypti in our dataset.
08.17.2020 - New paper in collaboration with HCPA published in Scientific Reports
Our paper Decreased expression of the thyroid hormone‑inactivating enzyme type 3 deiodinase is associated with lower survival rates in breast cancer, has been published in Scientific Reports. This work was developed in collaboration with researchers from the Endocrine Division of Hospital de Clínicas de Porto Alegre (HCPA) and found that a decreased expression of gene DIO3 is associated with lower survival rates in two populations of patients with breast cancer, from HCPA and from TCGA. In addition, we observed a hypermethylation of gene DIO3 in tumor samples, which may partially explain its lower expression values.
This paper is part of the PhD thesis of Dr Iuri Goemann, supervised by Profa. Dra. Ana Luiza Maia.
07.19.2020 - New preprint available: EPGAT, Gene Essentiality Prediction with Graph Attention Networks
The preprint EPGAT: Gene Essentiality Prediction With Graph Attention Networks, describing the results from the scientific initiation work of João Schapke (CIC/INF/UFRGS), in collaboration with Dr. Anderson Tavares, is now available!
The identification of essential genes/proteins is a critical step towards a better understanding of human biology and pathology. Computational approaches helped to mitigate experimental constraints by exploring machine learning (ML) methods and the correlation of essentiality with biological information, especially protein-protein interaction (PPI) networks, to predict essential genes. Nonetheless, their performance is still limited, as network-based centralities are not exclusive proxies of essentiality, and traditional ML methods are unable to learn from non-Euclidean domains such as graphs. Given these limitations, we proposed EPGAT, an approach for essentiality prediction based on Graph Attention Networks (GATs), which are attention-based Graph Neural Networks (GNNs) that operate on graph-structured data. Our model directly learns patterns of gene essentiality from PPI networks, integrating additional evidence from multiomics data encoded as node attributes. We benchmarked EPGAT for four organisms, including humans, accurately predicting gene essentiality with AUC score ranging from 0.78 to 0.97. Our model significantly outperformed network-based and shallow ML-based methods and achieved a very competitive performance against the state-of-the-art node2vec embedding method. Notably, EPGAT was the most robust approach in scenarios with limited and imbalanced training data. Thus, the proposed approach offers a powerful and effective way to identify essential genes and proteins.
07.06.2020 - Paper in collaboration with HCPA and INCA published in Cancers
The paper Calcium Signaling Alterations Caused by Epigenetic Mechanisms in Pancreatic Cancer: From Early Markers to Prognostic Impact, developed in a collaborative work among HCPA and INCA researchers was published in Cancers. In this work, we used a genome-wide DNA methylation approach to identify new pathways associated with Pancreatic ductal adenocarcinoma (PDAC) development. We observed that the Ca2+ signaling pathway had a high number of genes both significantly hypo- and hypermethylated in PDAC. Further analysis revealed the Calcium signaling pathway methylation alterations seemed to occur early in the carcinogenesis. Moreover, we showed that ORAI2, which controls Ca2+ influx through the plasmatic membrane, has an impact on prognosis, which suggests phenotypic importance of this finding. Therefore, our results bring the epigenetic dysregulation of the Ca+2 signaling pathway as a promising tool for PDAC management, either as diagnostic and prognostic biomarkers or as therapeutic targets.
This paper is part of the PhD thesis of Dra. Cleandra Gregório, supervised by Profa. Dra. Patricia Ashton-Prolla.
04.20.2020 - Paper accepted by Expert Systems with Applications
The paper describing the findings from the research project developed by Bernardo Trevizan as a scientific initiation student was accepted for publication by Expert Systems with Applications.
In this work, titled A comparative evaluation of aggregation methods for machine learning over vertically partitioned data, we performed an experimental comparison of previously proposed aggregation methods, including variations of elementary aggregators and meta-learning approaches, for a broad set of datasets in the domain of vertical data partitioning (when features are distributed among distint sources). As contributions to the research field we (i) provide evidence that none of the tested aggregation methods can consistently maintain its performance across all datasets, (ii) we experimentally show that performance is indeed influenced by the problems’ intrinsic characteristics (e.g., separability of classes and class imbalance), (ii) we identified relations between specific properties of datasets and the performance of aggregators, revealing characteristics that are more likely to have an impact on performance, and (iv) we created decision paths that can be used to visualize the relations revealed in the current work, as well as practical tools to guide the choice of the aggregation method in future works.
We acknowledge the support from FAPERGS (2017-2018) and CNPq (2018-2019), which provided the scientific initiation scholarships for this research.
01.29.2020 - Two papers published in Scientific Reports
2020 began with two papers published in Scientific Reports, describing the results of collaborations with researchers from HCPA and other institutions.
- A paper in collaboration with Dra. Thayne Kowalski and Profa. Dra. Fernanda Vianna from the Genomic Medicine Laboratory/HCPA, and other researchers from UFRGS and HCPA, that investigate the role of Cereblon-CRL4 complex in Thalidomide Embryopathy through bioinformatics and experimental approaches.
- A work describing a random forest model applied to peptidomics in the study of diabetic kidney disease biomarkers, led by Dra. Letícia Brondani and supervised by Profa. Dra Sandra Pinho Silveiro (HCPA).
01.26.2020 - Website updated and a summary of events in the last 1.5 years
After a long time without updates, the website was updated with information on current research projects and collaborations, latest publications (with a nice overview of published papers powered by Pubviz), and students supervisions. Summarizing the main events of the last 1.5 years:
- I had a son, Antônio. He was born on November/18 :-)
- Sheyla Paladini defended her dissertation in the Postgraduate Program in Cardiology/UFRGS, under my supervision. Part of her work was published by Molecular Diagnostic & Therapy.
- Ivaine Sartor concluded her PhD in the Postgraduate Program in Genetics and Molecular Biology/UFRGS, supervised by Profa. Dra. Patrícia Ashton-Prolla and co-supervised by me. Part of her results were published in Plos One.
- Laura Amaya Torres defended her dissertation in the Postgraduate Program in Computer Science/UFRGS, under supervision of Prof. Dr. Anderson Maciel and my co-supervision.
- Eduardo Soares de Abreu and João Pedro Hartmann Salomão completed their master degree in the Postgraduate Program in Computer Science/FURG. They were supervised by Prof. Dr. Adriano Werhli and Profa. Dra. Karina Machado, respectively, and co-supervised by me.
- A paper in collaboration with Igor Araujo, Profa Dra. Patricia Ahthon-Prolla, et al., was published in Gene
- Our paper about a systems biology approach to study gene expression regulators in pathological cardiac hypertrophy was published in Gene. It is the result of a collaboration with Profa. Dra. Andréia Biolo (HCPA/UFRGS) and Prof. Dr. Adriano Werhli (C3/FURG).
- Two papers in collaboration with Prof. Dr. Gustavo Machado (North Carolina State University) and researchers from University of Minnesota and University of Tasmania, were published in Scientific Reports and Journal of Animal Ecology
- Our work "Decreased expression of the Type 3 Deiodinase relates to lower survival in breast cancer", led by Prof. Dr. Iuri Goemann (HCPA) and supervised by Profa Dra. Ana Luiza Maia (HCPA), was presented during the XVII Latin America Thyroid Congress, and awarded the Merck Clinical Young Investigation Prize.
- I collaborated with the work "Synergistic effects between ADORA2A and DRD2 genes on anxiety disorders in children with ADHD", published in Progress in Neuro-Psychopharmacology and Biological Psychiatry, devleoped by researchers from UFSCPA, UFRGS, UNIVATES and UFPel.
- Since November/2019, I'm affiliated to the Postgraduate Program in Computer Science (PPGC), INF, UFRGS.
06.18.2018 - Two new publications in collaboration with the Endocrine Division/HCPA
Two papers developed in collaboration with researchers from the Endocrine Division of HCPA have been recently accepted for publication: "MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis" and "MicroRNAs and diabetic kidney disease: Systematic review and bioinformatic analysis.". These works are part of the PhD Thesis of Taís Assmann under the supervision of Dra. Daisy Cryspim, and count with the participation of researches from Instituto da Criança com Diabetes e do Hospital Conceição.
10.06.2017 - Paper in collaboration with Lab. of Human Pancreatic Islet Biology/HCPA accepted by Endocrine Connections
A paper written in collaboration with MSc Taís Assmann, Dra. Daisy Crispim, and Dra. Bianca M. Souza, from the Laboratory of Human Pancreatic Islet Biology, Endocrine Division of HCPA, was recently accepted for publication in Endocrine Connections. The paper is titled "MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatics analysis", and has identified eigth microRNAs (miR-21-5p, miR-146a-5p, miR-148a-3p, miR-181a-5p, miR-210-5p, miR-342-3p, miR-375 and miR-1275) related to immune system function, cell survival, cell proliferation, and insulin biosynthesis and secretion with consistent dysregulation in Type 1 Diabetes Mellitus patients, representing potential circulating biomarkers of this disease.
09.14.2017 - Talk on 'Big data and Bioinformatics' presented at the 37a Semana Científica/HCPA
At September 14th, I had the pleasure to participate as invited speaker, together with Prof. Dr. Maria Yury Ichihara (CIDACS/FIOCRUZ) and Eduardo Cipriani (IBM), in the Roundtable "Exploration of Large Databases", organized by Prof. Suzi Camey as part of the 37a Semana Científica of HCPA (37th Scientific Week of Clinical Hospital of Porto Alegre). My talk was focused on the opportunities and perspectives for Bioinformatics in the scientific activities of HCPA.

12.10.2016 - Best Poster Award received at ISCB-LA 2016
Our work presented at the 4th International Society for Computational Biology Latin America Bioinformatics Conference (ISCB-LA 2016), titled "Differential network analysis for the identification of common and specific regulatory mechanisms between ischemic and idiopathic dilated cardiomyopathy", has received the 1st Place in the Best Poster MST Awards, sponsored by the Medical Science & Technology Journal. This work is developed in collaboration with Daniel Sturza Caetano, Dra. Andreia Biolo, Dra. Nadine Clausell, and Dr. Luis E. Rohde.
12.04.2016 - Research grant proposal approved by FAPERGS
At November 23th, FAPERGS (Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul - State Research Funding Agency of Rio Grande do Sul) disclosed the final results of the PRONUPEQ 2016 (Programa de Nucleação de Grupos de Pesquisa - Nucleation Program of Research Groups) call to fund research projects in line with the priority areas defined by the Secretary of Economic Development, Science and Technology of the State of Rio Grande do Sul. Our project titled "Bioinformática aplicada à medicina personalizada: do diagnóstico ao tratamento de doenças" ("Bioinformatics applied to personalized medicine: from diagnostic to diseases treatment") was one of the nine research projects selected for funding! This project was written in close collaboration with Prof. Marcio Dorn (INF/UFRGS, Project Coordinator) and aims to bring together specialists in the areas of Computer Science, Medicine, Genetics and Biology to tackle important challenges related to drug development and personalized health care. Specifically, together with professors and researchers from Institute of Informatics and Biotechnology Center of UFRGS, from Hospital de Clínicas de Porto Alegre (HCPA), from Universidade Federal do Rio Grande (FURG), as well as from other collaborating institutions, we aim to develop and apply bioinformatics methods to find new potential biomarkers and therapeutic targets for cardiovascular diseases and cancer (especially hereditary cancer). The list of researchers and main collaborators of the project is given below:
Universidade Federal do Rio Grande do Sul (UFRGS)- - Prof. Dr. Marcio Dorn (INF/UFRGS)
- - Profa. Dra. Mariana Recamonde Mendoza (INF/UFRGS)
- - Profa. Dra. Ana Bazzan (INF/UFRGS)
- - Prof. Dr. Hugo Verli (CBiot/UFRGS)
- - Prof Dr. Guido Lenz (CBiot/UFRGS)
- - Dr. Rodrigo Ligabue Braun (CBiot/UFRGS)
- - Profa. Dra. Karina dos Santos Machado (C3/FURG)
- - Prof. Dr. Adriano Werhli (C3/FURG)
- - Profa. Dra. Andreia Biolo (Serviço de Cardiologia/HCPA)
- - Dr. Michael Andrades (Centro de Pesquisa Experimental/HCPA)
- - Profa. Dra. Nadine Clausell (Serviço de Cardiologia/HCPA)
- - Prof. Dr. Luis Eduardo P. Rohde (Serviço de Cardiologia/HCPA)
- - Profa. Dra. Patricia Ashton-Prolla (Serviço de Genética Médica/HCPA)
10.15.2016 - Abstract accepted for oral and poster presentation at ISCB-LA 2016
The abstract titled "Differential network analysis for the identification of common and specific regulatory mechanisms between ischemic and idiopathic dilated cardiomyopathy" has been accepted for both oral and poster presentation at the 4th International Society for Computational Biology Latin America Bioinformatics Conference (ISCB-LA 2016), which will be held at Buenos Aires, Argentina, on November 21-23, 2016. This work is being developed in collaboration with Dra. Andreia Biolo, Dra. Nadine Clausell and Dr. Luis E. Rohde, medical doctors at Hospital de Clinicas de Porto Alegre and professors of the Post-Graduate Program in Cardiology and Cardiovascular Sciences of Universidade Federal do Rio Grande do Sul (UFRGS), and with Daniel Sturza Caetano, an undergraduate student in Biomedicine (UFRGS).
08.04.2016 - Course: Bioinformatics for Health Science
In collaboration with Profa. Dra. Ursula Matte (Genetics Department of UFRGS), her graduate student Delva Leão, and Profa. Dra. Silvia Olabarriaga (AMC e-Science), we are organizing and extension course on "Bioinformatics for Health Science" that will be held on August 22-26, at the Experimental Research Center of Hospital de Clínicas de Porto Alegre. The aim is to introduce concepts and tools of Bioinformatics that may be helpful in projects related to health sciences, and to estimulate their application in research projects under development by undergraduate students, graduate students, postdocs, and professionals. Topics covered by the course include: new generation sequencing, genetic variant analysis, differential expression analysis, functional enrichment and systems biology, Registrations can be done here. The course will be taught in portuguese. More information below (click on the image to enlarge):
07.01.2016 - Started an Associate Professor position at INF-UFRGS
On June 29th, I started a tenure-track Associate Professor position ("Professor Adjunto") at the Applied Informatics Department of the Institute of Informatics, Federal University of Rio Grande do Sul (UFRGS). UFRGS is the largest public university in the south of Brazil and was ranked as the best university in Brazil in an assessment made by the Ministry of Education. Moreover, the Institute of Informatics is one of the largest Computer Science departments, offering two undergraduate degrees (Computer Science and Computer Engineering) that are among the best in the country, and a Postgraduate Program in Computing that figures among the top 5 programs in Brazil.
04.22.2016 - Paper in collaboration with Profa. Luciana Tovo-Rodrigues accepted by Neuropsychiatric Genetics
A paper developed in collaboration with Profa. Dra. Luciana Tovo-Rodrigues (UFPel) and researchers from the Genetics Department of UFRGS (Profa. Dra. Mara H Hutz, Estela M Bruxel, Jaqueline B. Schuch, Deise C. Friedrich), Federal University of Bahia (Profa. Dra. Vanessa Rodrigues Paixão-Côrtes) and Hospital de Clínicas de Porto Alegre (Prof. Dr. Luis Augusto Rohde) has been accepted for publication in the American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. The paper is titled "The role of protein disorder in Major Psychiatric disorders" and can be accessed here.
04.01.2016 - Concluded Postdoc at HCPA
In March 2016 I concluded by Postdoctorate in the Experimental and Molecular Cardiovascular Laboratory of Hospital de Clínicas de Porto Alegre. My research was focused on the application of Bioinformatics and Systems Biology tools to uncover the action of transcriptional and post-transcriptional mechanisms in the regulation of gene expression contributing to the development of cardiovascular dysfunction and diseases. I'm also a collaborator of several on-going research projects carried out by Laboratório de Medicina Genômica (Experimental Research Center/HCPA) and Laboratory of Human Pancreatic Islet Biology (Endocrine Division/HCPA), helping with the analysis of large-scale genomic data and investigation of miRNAs' functional role in diseases such as Cancer and Type 1 Diabetes Mellitus.


